A new hierarchical classification methodology
As part of my recent paper scPRINT I stumbled upon an issue leading me to build a new kind of classifier for hierarchical-tree-based labels often defined in ontologies. What am I talking about?
A big issue plaguing the field of cell biology and some others is the lack of common terms to talk about things, like diseases, cells and tissues. Fortunately we now have really good ontologies that define and relate these terms in hierarchical trees: e.g. CL
But how do we make the best use of these for a classifier? In the cellxgene datasets, labels of a class come in different precisions, e.g. you might get “neurons” in one dataset, and other datasets you will get “dopaminergic neuron”; “GABA neurons”,…
With my hierarchical classifier, I define a training procedure on a classifier which only predicts the leaf nodes of the ontological tree (the most precise values possible). If the annotated label is something imprecise like “neurons”, the model will still compute a loss on its precise prediction by considering each children labels of neurons as a positive label.
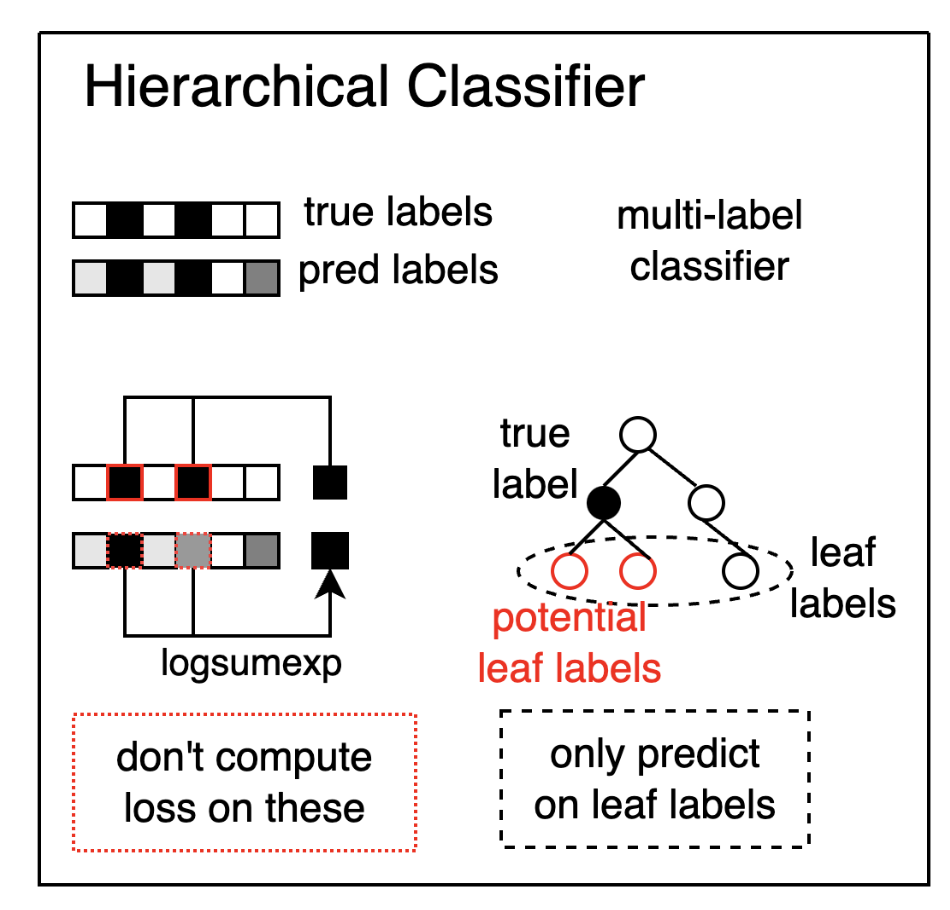
Lastly note that the training procedure doesn’t just put each children labels to “True” but has to compute the max() over the children labels so as to enforce that the model only selects one label for which it has the most certainty or attribute high certainty to all labels if it doesn’t know which one to choose.
People can use tools like lamindb to convert from raw annotations to ontological annotations based on fuzzy matching. They can also use scPRINT to relabel the cells in datasets. For now labels such as cell type, disease, ethnicity, sequencer, sex are available. I will soon add Age and Tissues prediction to this mix.
Some work remain to be done on scPRINT to use its prediction uncertainty to better help labelling of cells and aggregate cell level labels to generate more consistent annotations. Help and PRs are very welcomed: scPRINT
Leave a comment