Managing GRNs and What They Mean
Managing GRNs and What They Mean
In my recent paper scPRINT, I went into a deep dive on gene regulatory networks. Many findings are in the manuscript but I had some additional thoughts to share and wanted to present a tool I built to work with GRNs.
What are Gene Networks?
First we don’t use the term GRN most of the time but Gene Networks. This is because when GRNs are mentioned, they are almost always restricted to TF-gene connections. But gene expression / regulation is way more complex than this and driven by many more elements than just TFs. ♾️
Indeed, regulation happens with TF but also co-factors, cohesins and other proteins transporting, degrading, splicing the RNAs, shaping the DNA and working with TFs. Not only that but it is also well known that TFs themselves can be influenced and activated by pathways comprising many genes less related to transcription regulation. 🦠
Finally, a new realization in biology is that regulation also happens through RNAs interacting with each others, and with the DNA. So much happening but we are fixated on TF-genes… 🔍
The Problem with Simple GRNs
The big issue in my opinion is when these simple computationally inferred GRN are used to perform simulation of gene expression or are conflated with a model of the cell! 😲
The cell is very complex and TF-gene binary connections won’t cut it, by a mile. ⛔ This is to me a big problem in the GRN and GRN inference field. We will need to better define what GRNs are and maybe use another term to talk about the TF-gene constrained networks: like TFGN instead of GRN.
Given all this information -and the still unknown amount of non coding RNAs in human cells- we have to accept that a true Gene Regulatory Network should implicate all RNAs, non coding RNAs, DNA regions, proteins, and likely other molecular products. 👩👩👧👦
Finally, connections are not binary, they are not weighted, molecular interactions are complex, can create complexes, partially interact and have conditionals. We will also need better representation of connections, likely using embedding representation of vertices. 🔀
Where Do We Go From Here?
1. Theoretical Foundations 📋
We need good theoretical definitions that most people agree on, however murky things still are to us.
2. Better Tools and Infrastructure 🧰
We need to build tools, visualisation, data-structures, and benchmarks that represent these definitions and concepts and help us interrogate the current and future datasets we might generate. This is what I tried to start with GRnnData and benGRN. But lot more work is needed.
3. Advanced Sequencing Methods 📏
We need better sequencing methods than mRNA sequencing. Single cell is only a first step but we need to measure non coding and rare RNAs with much higher precision.
Who knows, it might be that a large-scale, high-depth, single-cell total-RNA-sequencing methodology with correct molecular amplification / reduction methods is all that we need to understand regulation… 🤔
GRnnData
in scRNAseq data we have been fortunate to get many great python toolkits like anndata and scanpy, part of the scverse suite of tools. Allowing us to work together with a common set of standards. 📜
However, working with gene network methods, I have seen various ways to store them throughout the different papers and benchmarks. Often as some kind of tsv/csv/… file with some kind of a gene-gene list. This lack of standard made it quite hard for me and other members of the lab to work with gene networks 🙉
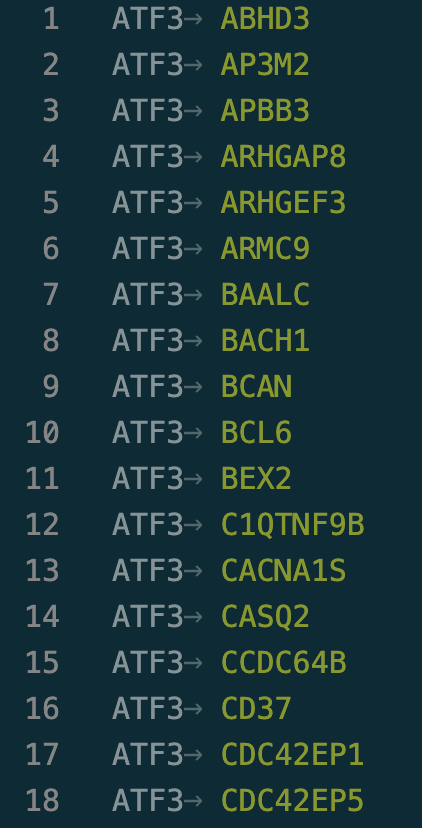
Interestingly, there is a possible standard for it! 🎉AnnData contains the .varp field which is made to store var to var (e.g. genes to gene) relationships. However not many people use it…
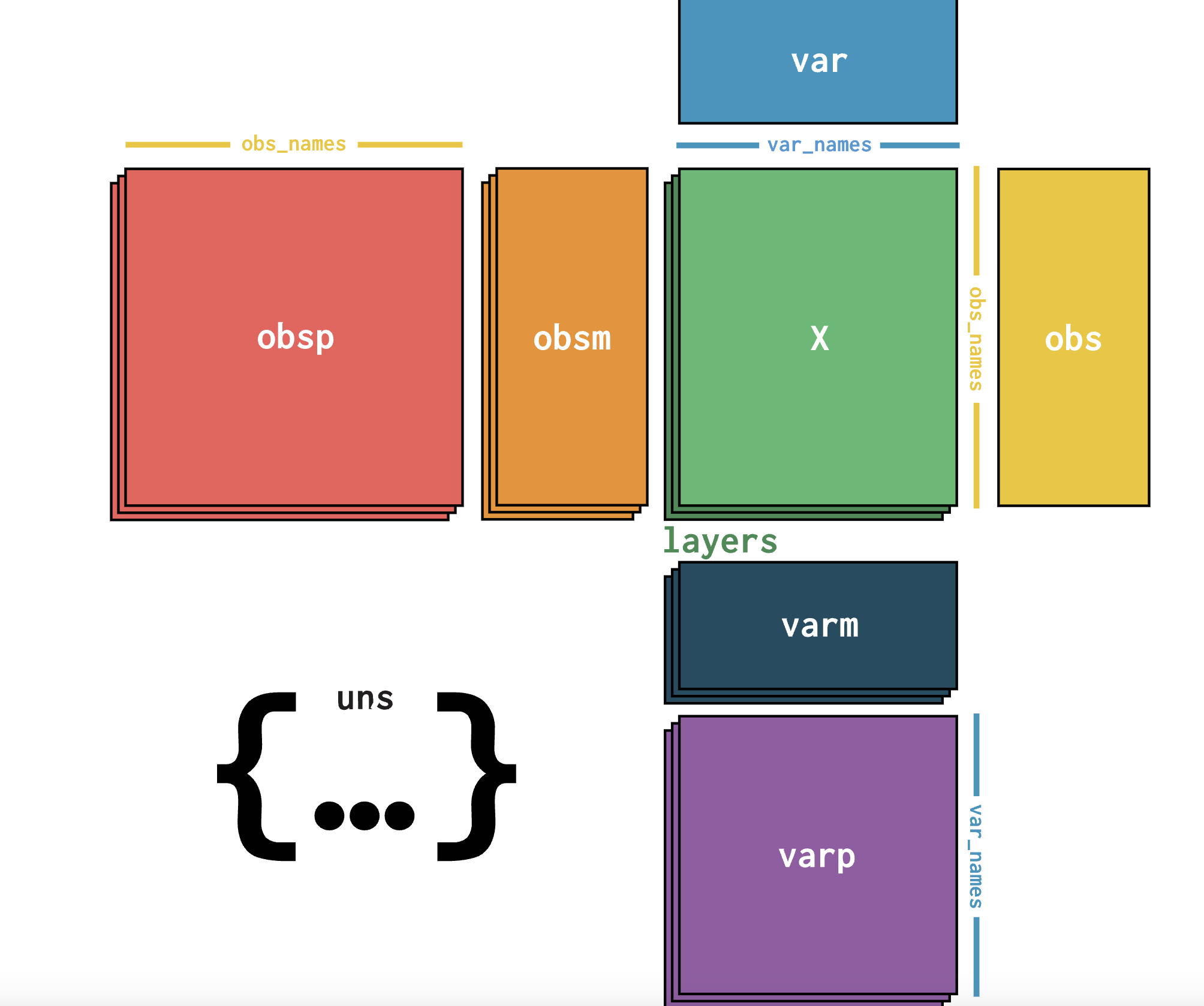
I thus decided to formalize this usage by creating GRnnData: it is basically a way to important many different gene-gene format to an AnnData file. 💁 But it also contains more bells and whistles to work with gene networks like subsetting the network to some genes, extracting targets, and plotting the networks 💹.
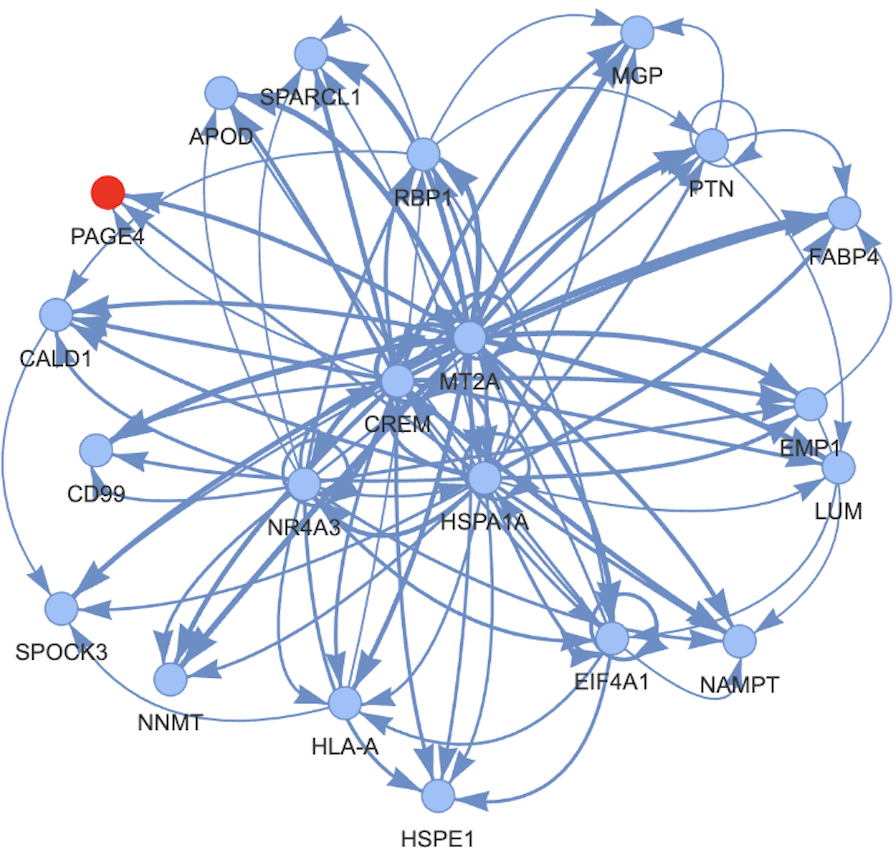
But GRnnData can do more and integrates some utils functions doing things like clustering, centrality measures, enrichment and more. Go check [GRnnData]https://github.com/cantinilab/GRnnData if you work with gene networks!

Leave a comment