About the AIVC paper
Reading the AIVC manuscript further, I felt like adding some more comments on this topic, which I did before on X too.
Being able to model a cell with large amount of multi modal data and multi scale AI models is something I have talked about before on X and in my recent manuscript
To my mind there is two questions:
- Do we have the right algorithm and compute?
- Do we have the right data and scale?
For the first part, while being just a guess, seeing the abilities of recent AI models and the scale of their deployment in both language, vision, and structural biology. It stands to reason that we might be able to model a cell with some $\epsilon$-accuracy with current software and hardware, e.g. 10^13 params model trained and doing inference on 10,000 GPUs.
For the second part it is harder to be as optimistic. A human body is comprised of 100 trillion cells and other microbes. All shaped by their environment. The cell doesn’t, in fact, follow the simple diagram presented in the AIVC paper:
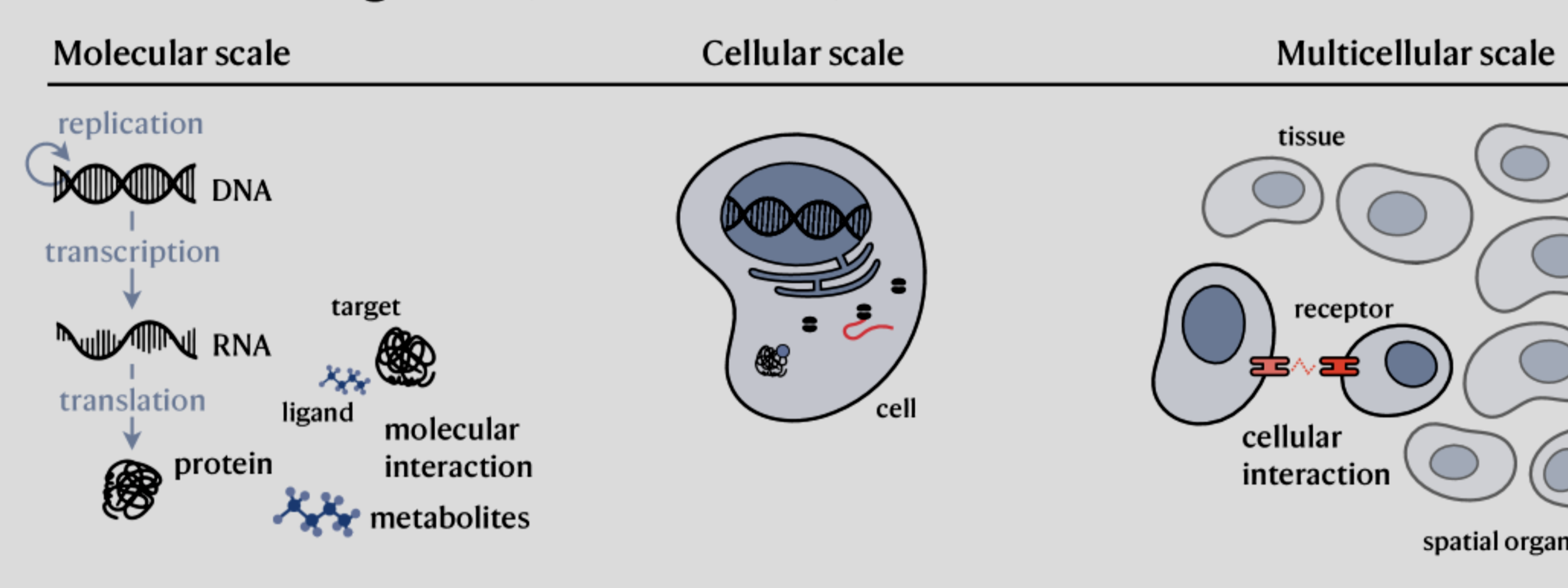
But likely this diagram:
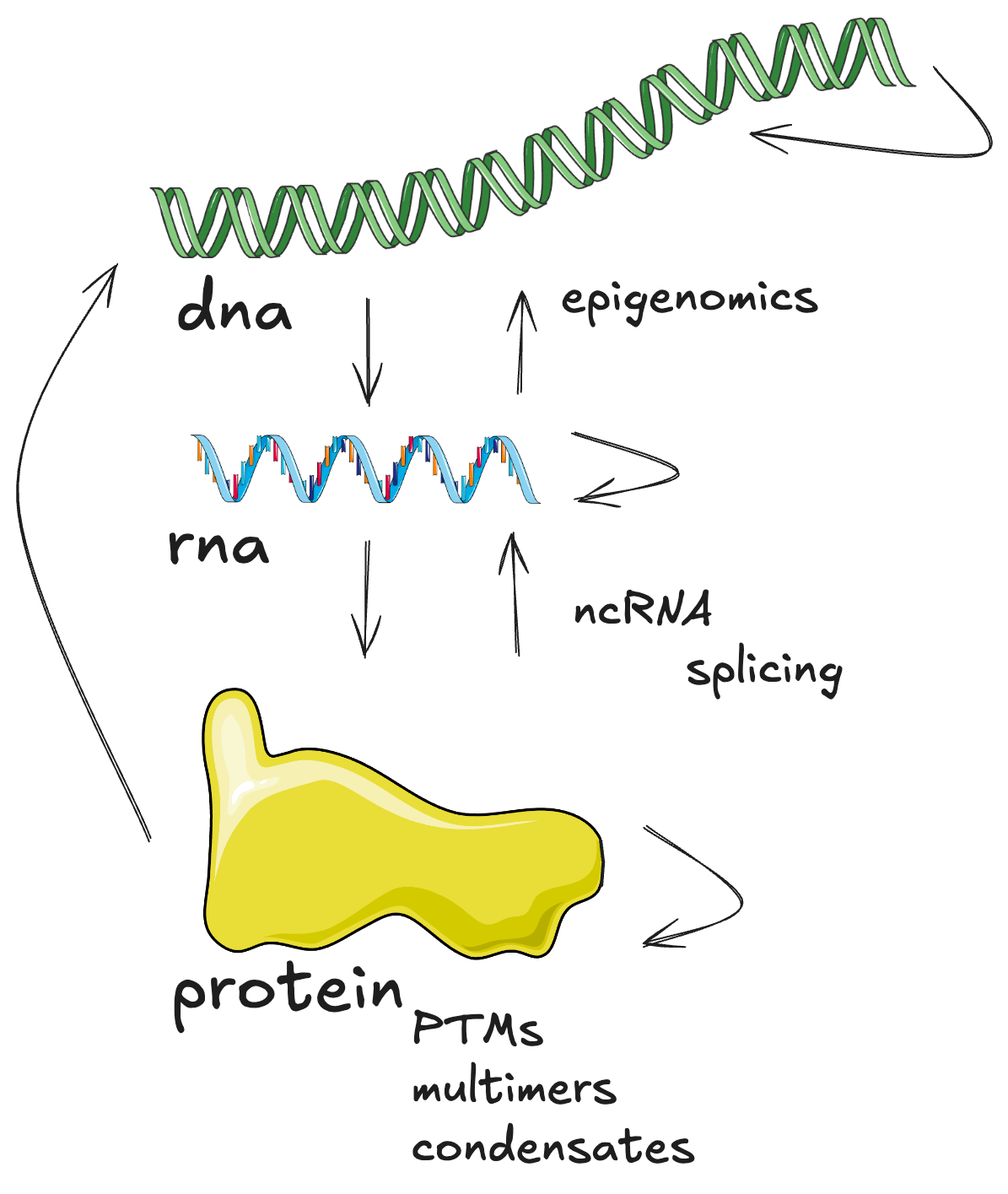
Proteins are not RNAs and not genes. While genes can stay the same many RNAs can come out. Same with proteins. And vice versa. Thus one cannot encode it all in one representation. They are different objects.
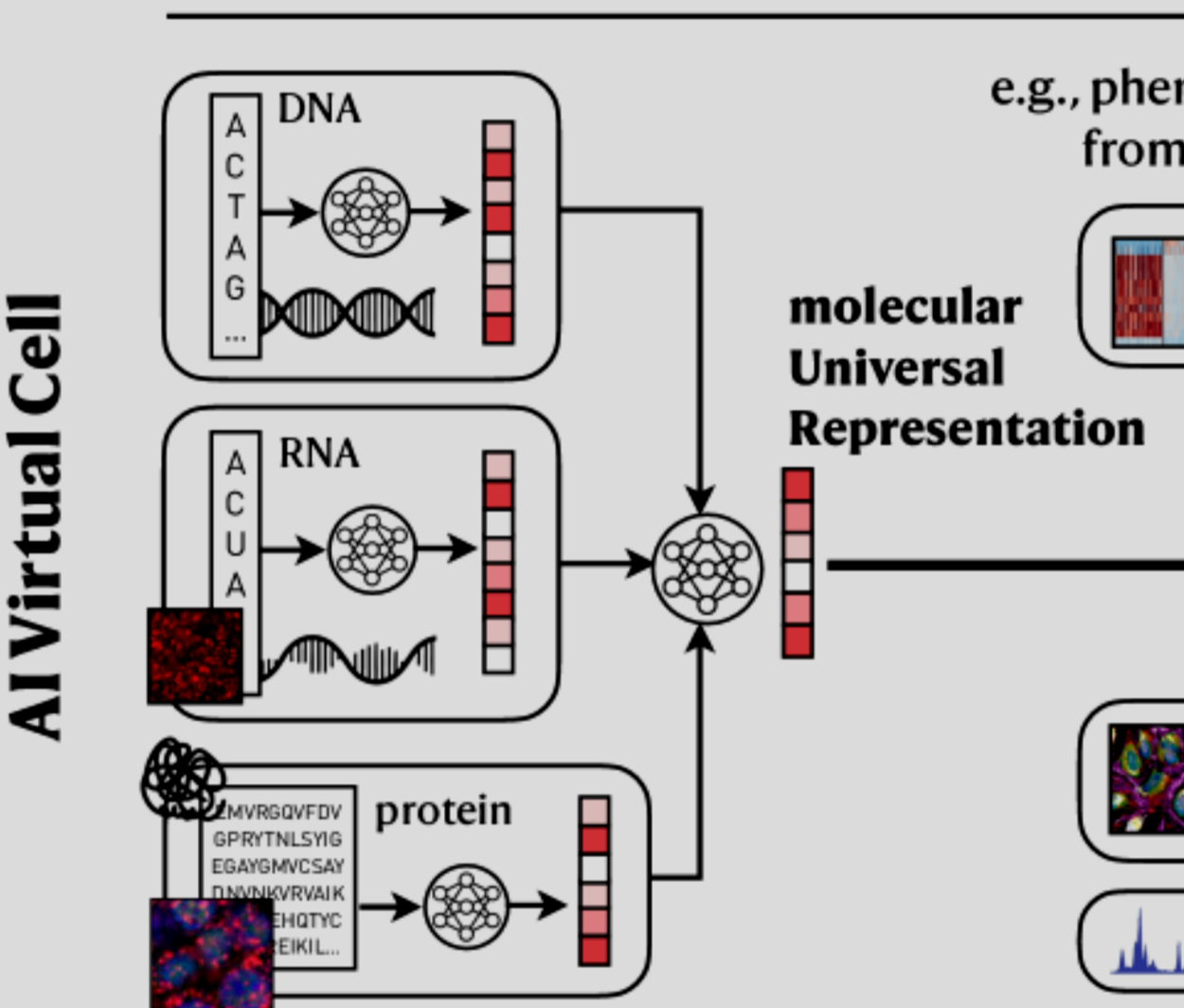
Is it just a modelling problem? No, and these apparent simplifications are not there by mistake. They are due to our measurement abilities or actually, inabilities. We cannot measure well these diverse proteins / RNAs nor their interactions…. yet #ExSeq, #DNApaint, …
We can use AI to model proteins indeed but we knew everything about the theory of proteins, their elements and could measure and model their structures for decades prior.
So to me the future of AIVC is not really in the hands of the AI community but in the hands of the bio community. How do we measure these things better so as to have at least a good picture of how they work.

Leave a comment